
Sending flu packaging

What makes influenza so difficult to control is the way in which it evolves. All viruses evolve, with minor genetic mutations either being carried forward to new generations or dying out. This normal form of evolution in the influenza viruses which circulate in humans is called antigenic drift: the virus slowly changes over time, and we retain some partial immunity to the evolved virus thanks to our immune systems remembering similar past infections. However, things become very dangerous when antigenic shift occurs. In this case two different strains of influenza mix together to create what is essentially a new disease to which we have no immunity.
During influenza infection, viruses spread from cell to cell, hijacking cellular machinery to make more copies of themselves. The influenza genome is unusual in being made of several segments, rather than one continuous genome. The different segments are replicated separately, but must end up together inside a new virus particle. There are eight segments in the most common form of influenza, and all contain important genes, so a virus needs a full set of segments to be fully viable.
The Disease Dynamics Research Group at the University of Cambridge is interested in how genome segments are brought together to make the new virus particle: a process known as packaging. Why is this important? Being segmented it is easy for influenza to swap genes between different strains: a process called reassortment. Reassortment is the mechanism behind antigenic shift and was implicated in all the 20th century flu pandemics. The 2009 swine flu virus is believed to be a multiple reassortant, where several past reassortment events led to the virus that could spread across the globe.
Let me count the ways...
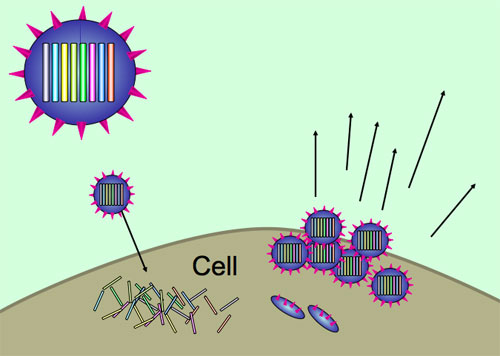
Viruses use host cells to replicate themselves, with each new virus particle (virion) containing all eight segments of the influenza genome.
Does influenza package segments at random or is there a special mechanism to ensure each virus particle gets the eight different segments? If we number the segments 1 to 8, then random packaging is equivalent to picking eight numbers at random from this set, say {5, 5, 7, 4, 1, 6, 3, 8}. But this example won't do, as it doesn't contain all eight segments. The number of ways of picking exactly one of each of the segments is
8 x 7 x 6 x 5 x 4 x 3 x 2 x 1 = 8! = 40,320
So the chances of picking a complete set at random is 8!/88 = 0.0024, or less than 1 in 400. Random packaging clearly isn’t a good strategy!
Looking for signals
Current experimental evidence suggests that segments are indeed packaged specifically. But for packaging to be specific, there must be something about each of the eight segments that acts to distinguish it from the others: a label or packaging signal. But how do we find these signals? Here is where a mathematical and computational approach helps.
It appears that the packaging signals are embedded in regions of the genome that encode proteins, like a message hidden within another message. There are many possible sequences that encode the same protein, and the virus ought to be able to mutate to any of these synonymous sequences. But in parts of each segment of the virus, the genome sequence does not vary despite many variations being possible. This seemingly inexplicable constraint may indicate that these regions act as packaging signals.
Our computational techniques make use of the large amount of influenza genome sequences that are publicly available. The techniques allow us to align hundreds of examples of the genome sequence and come up with a score for every position in the sequence (based on the number of variations possible, as well as some other factors) where low scores mean there's suspiciously little variation, when variation would have been possible.
Once we had pin-pointed suspicious regions in the genome colleagues at the Department of Pathology explored them further in the lab by engineering mutant viruses to see if the packaging process is broken by changes in those locations in the genome. It seems that our methods are identifying regions of interest. Our other work has included applying our approach to explore rotavirus (which causes diarrhoeal disease and is a major cause of death in young children worldwide), which also has a segmented genome. Current research is to extend our methods to look for interesting regions in the HIV genome, in collaboration with the Department of Medicine.
Understanding the packaging process of influenza and uncovering the packaging signals that drive it would be a major step in understanding how viruses work. Not only is this of basic virological interest, it could also lead to possible treatments. And importantly, it would lead to a better understanding of the reassortment of "normal" flu with avian or swine flu, that allow dangerous viruses to become adept at infecting humans. Perhaps this research will give us another defence against pandemics in the future.
This is an edited version of the article Influenza virus: it’s all in the packaging by Dr Julia Gog, Lecturer in Applied Mathematics and Head of the Disease Dynamics Research Group at the University of Cambridge. You can read the full article here, or find out more about mathematics and infectious diseases in this collection of Plus articles.