Revolutionising the power of blood tests using AI
When your doctor orders you to have a blood test, chances are that the lab will perform a full blood count. This test gives information about the kinds and numbers of cells in your blood. It's a good indicator of your general health and gives clues about specific health problems, such as iron deficiency or vitamin B12 anaemia.
When a full blood count is done only 20 of thousands of measurements are recorded.
Full blood counts are performed in huge numbers. Around 150 million a year in the UK alone, rising to 3.6 billion globally. Each time 20 cell-level measurements from the blood are recorded in a patient's file. However, the machine performing the test, called a haematology analyser, takes thousands of measurements from a patient's blood that aren't usually recorded.
This data presents a vast, untapped goldmine of information. It could provide an early warning sign of outbreaks of infectious diseases, and it could also be used to screen for people at high risk of particular diseases. Since the data is already there, routinely measured as part of people's full blood counts, this would provide a cheap and effective way of improving public and individual health.
The reason this isn't already happening is that it's humanly impossible to analyse the vast amount of data to find the necessary clues. Enter machine learning, a form of artificial intelligence that has generated a lot of attention over recent years. It's used routinely to help you with your online shopping or communicate with a computer by voice, and it's behind applications such as ChatGPT and Bard. Machine learning involves algorithms learning to spot patterns in data that humans usually aren't able to see, and making predictions based on these patterns — you can see a quick introduction in this article, and a more detailed explanation in this article.
Blood counts
A project called BloodCounts! is now busy directing the power of machine learning at the rich data that come from full blood count tests. "From a clinical perspective it's a no-brainer to use this massive amount of data," says Nick Gleadall, Assistant Professor in Clinical Genomics of Transfusion and Transplant at the University of Cambridge and member of BloodCounts!.
BloodCounts! is an international consortium of mathematicians, clinicians and data scientists led by Carola-Bibiane Schönlieb, Professor of Applied Mathematics at the University of Cambridge. Member organisations include health authorities and universities in the UK, the Netherlands, Belgium, Ghana, The Gambia, India and Singapore. However, the methods that are being developed are ultimately intended for even wider international use. In particular, countries with a less developed public health infrastructure could benefit. "The full blood count is such a basic test that’s done anyway as part of normal medical care," says Gleadall. "If you remember the pandemic, even rich Western countries had trouble rolling out PCR testing. Some countries just don't have the option to scale up in this way. Using data that already exists can be of huge benefit here."
BloodCounts! isn't the only project gearing up to using machine learning in medicine and health care. Researchers are exploring a range of other uses, from helping with breast cancer screening to diagnosing Alzheimer's disease. Michael Roberts, Senior Research Associate of Applied Mathematics at the University of Cambridge and a member of BloodCounts!, says there's no doubt it will soon become an integral part of medicine and healthcare. "It's cheap, it's massively scalable, it never gets tired, it doesn't work shifts, and it'll give you the same result every time for the same data," he says.
Spotting patients at risk
There are, however, a range of challenges that need to be overcome first. The research done by BloodCounts! falls into two strands. One is to develop algorithms that can identify people at high risk of a particular disease, for example leukaemia, kidney cancer, and pregnant women at risk from preeclampsia.
These algorithms are trained on existing data that comes from patients we know had the disease in question, or went on to develop it, when their blood was tested. From this training data the algorithms learn the patterns in blood count data that are linked to the disease. Once the training is complete, the algorithms can then be used to analyse blood counts from new patients and raise the alarm if a disease related pattern is present. This type of machine learning is called supervised learning because the algorithm learns from data for which the answer (whether someone had the disease or not) is already known– you can find out more here.
Spotting outbreaks
The main strand of BloodCounts!, however, is about pandemic surveillance: developing algorithms that can spot the outbreak of a disease in a particular region from the routine analysis of blood counts from people living there. Excitingly, the methods that BloodCounts! are developing will even be able to spot outbreaks of diseases that are completely new.
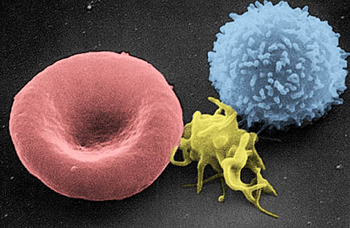
Artificially coloured electron micrograph of blood cells. From left to right: erythrocyte, thrombocyte, leukocyte.
In this case, there is no data from people with the disease for the algorithms to be trained on, which is why something called unsupervised learning needs to be used. A clever example of the kind of thing Roberts and his mathematical colleagues are working on is similar to algorithms that can be used to compress images so they use up less memory on your computer. Initially, an image is stored as an array of numbers, one number for each pixel giving the pixel's colour. Machine learning algorithms, called autoencoders, can learn how to draw from this number array the most important features of an image and how to reconstruct the image from those features. This way, it's only the information describing the features that the computer needs to store.
In a similar way, an algorithm can learn to compress a person's full blood count, reducing the thousands of numbers down to the thirty of so that best represent the it, and how to reconstruct the full blood count from those thirty numbers. If the algorithm is then confronted with an unusual blood count, say from a patient who has a new disease, this will be noticed.
"If I took all this year's full blood counts, the algorithm could reasonably and robustly compress and reconstruct them," explains Gleadall. "So when the algorithm sees [unusual] blood counts that it has never seen before come through, it falls apart. It doesn't know what to do. You can measure that and say 'hey, what's going on with this group of samples?'" Back in 2021 the BloodCounts! team developed machine learning methods based on this idea and showed that it would have been able detect a COVID-19 outbreak in Cambridge. The challenge now is to make this work for larger populations and different cohorts of people.
Shifting domains
This latter challenge touches on one of the main reasons why machine learning isn't already being used routinely in healthcare and medicine: when you have trained an algorithm on one data set, say blood counts from a particular hospital, there's no guarantee that it'll give you correct answers on another data set, such as blood counts from another hospital.
The reason is that data collected in a particular place can be biassed. "All of the factors of clinical practice can influence a blood sample," says Roberts. "How long a sample is left out after it comes out of your arm, the temperature of the day, the person who is dealing with the sample — all that can influence the measurement. Then there's the machine reading the sample. Different machines are calibrated differently. On different days they perform differently, and just after they've been serviced they perform differently. You can detect these differences in the data, but for machine learning this is terrible."
This problem of domain shift, where the data set an algorithm is trained on can differ from the data sets it's going to be deployed on, is something people have long been familiar with, for example when it comes to using machine learning to assess medical images. "For example, chest X-rays always look different depending on what scanner they're done on and what hospital they're done at," says Roberts. "Also there may be embedded text in the image, which says, for example, whether a patient has been lying down or standing up. A patient lying down is probably sicker, so the algorithm will learn to predict that the lying down patient will be worse off. So the algorithm is learning all these shortcuts that you really don't want it to learn."
Luckily, it's possible to develop algorithms that ignore patterns associated with a specific machine, or setting, and only learn the important information that is truly relevant. This is called domain generalisation. "We have some really nice recent results, where you can see that as you add more machines to your network the quality of performance massively increases. But if you don't use this domain generalisation technique the quality really falls off."
Federated learning
Getting your algorithm to make correct predictions is obviously paramount in all applications of machine learning, but applying machine learning in medical settings comes with an extra challenge: you're dealing with intimate medical information about real people who have a right to privacy. To guarantee this, you want to minimise the movement of data from place to place. "Previous studies on medical imaging and clinical data have always been biassed because you could only involve a single centre with a small amount of data or a couple of centres that are willing to work together," says Roberts.

An illustration of federated learning where an algorithm is shared between a central server and various local sites. Image: MarcT0K, CC BY-SA 4.0.
Here too there's an algorithmic solution the BloodCounts! team have been helping to develop further, called federated learning. "The idea is that you leave the data at the site it resides and you share the algorithm between the sites. The algorithm gets trained at each site and then gets shared back. Throughout this whole process the privacy of the data is retained."
Machine learning algorithms learn by repeatedly tuning internal parameters, also called weights, until the error in the predictions they make is as small as possible. In federated learning it is only these weights, not the data used for training, that is shared by individual sites with the central site. Another clever algorithm at the central site then uses a sort of averaging process to aggregate the information from all the sites' weights in the algorithm, and sends the updated algorithm back to the sites. These then tune the weights further using their training data, send the new weights back to the central site, and so on. "Slowly you get this better and better algorithm because it has all the knowledge of all the sites that took part," says Roberts.
Beyond maths
Federated learning and domain generalisation are really entire research areas of their own, but the BloodCounts! team is working to overcome most of the important mathematical hurdles. However, with a project of this size and ambition, lots of other things need to be put in place too. The three main UK hospitals that are taking part — University College Hospital London, St Bartholomew's in London, and Addenbrooke's in Cambridge — are technologically advanced and have large data bases, but bringing all this data into a standardised format and setting up the necessary infrastructure and computational environment involves a lot of work and bureaucracy.
The team also had to submit an extensive application to the Research Ethics Committee, which involved engaging with patients and members of the public. "We had positive feedback from real patients," says Roberts. "The way we structured our secure research environments and all the technology to keep data safe has convinced them that this is a transformational study which holds great promise."
"Most people say, 'if you can make better use of my data, then why not?'", says Gleadall. "But informed consent is a big issue with data at this scale, so it was important to us to do the patient and public involvement work as part of our ethics application."
All in all Gleadall and Roberts think it'll be a few more years until BloodCounts! is fully operational, routinely scanning full blood counts. When it's live it'll be one of the largest-scale applications of machine learning in medicine and healthcare yet. With all the hurdles that needed to be overcome — involving maths, technology, data science, infrastructure, and ethics — it's likely to point the way for other uses of artificial intelligence in this field.
About this article

Nick Gleadall.
Nick Gleadall received his PhD in 2020 from the University of Cambridge. He is currently Assistant Professor at the University of Cambridge and has a focus in genomics, in particular the use of high-throughput genotyping or sequencing technologies and large datasets in transfusion medicine. Prior to his PhD he has an establish history of translating cutting edge research into routine NHS service.

Michael Roberts.
Michael Roberts is is a Senior Research Associate of Applied Mathematics at the Department of Applied Mathematics and Theoretical Physics and at the Department of Medicine. He is a member of the Cambridge Image Analysis group (CIA) and leads the algorithm development team for the global COVID-19 AIX-COVNET collaboration.
Marianne Freiberger is Editor of Plus. She interviewed Gleadall and Roberts in April 2023.